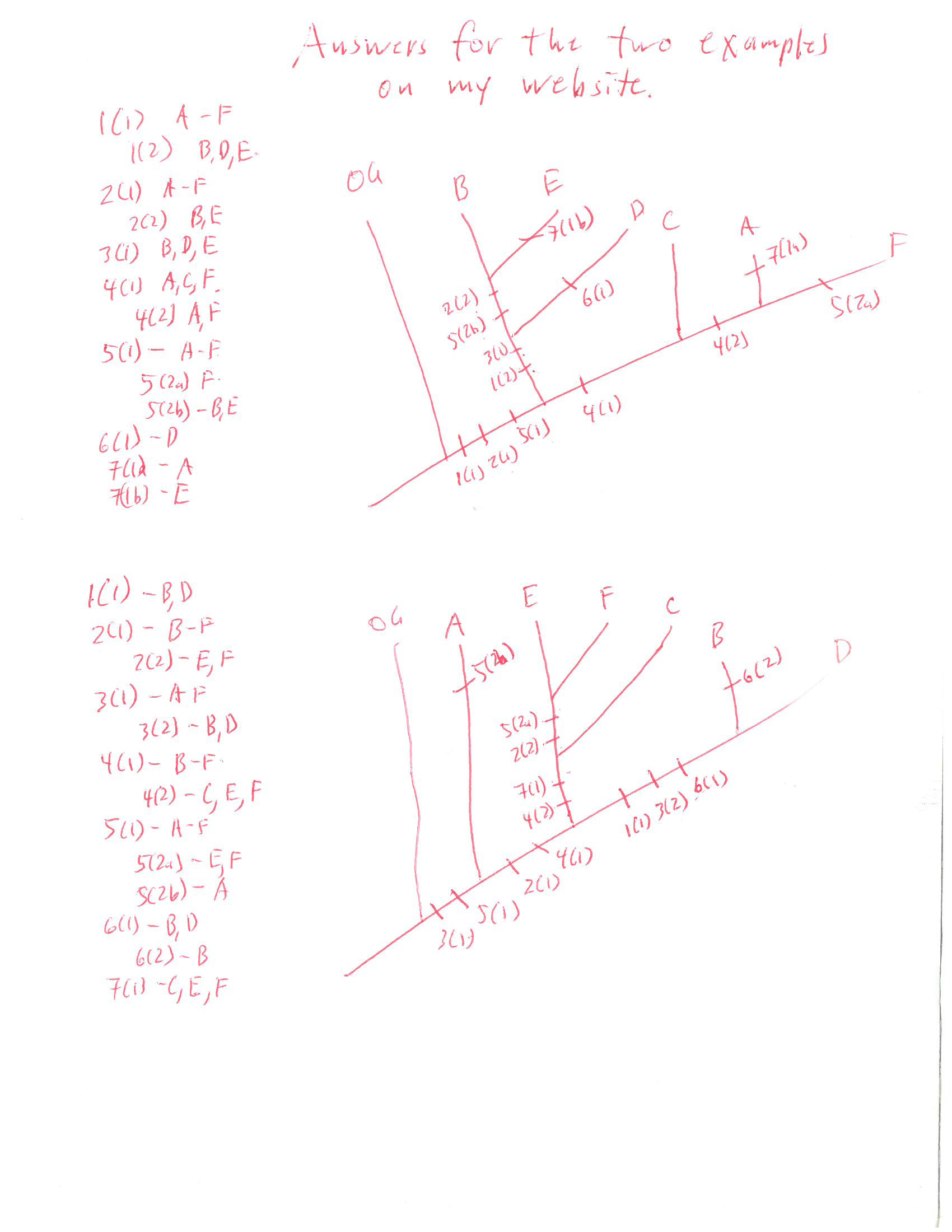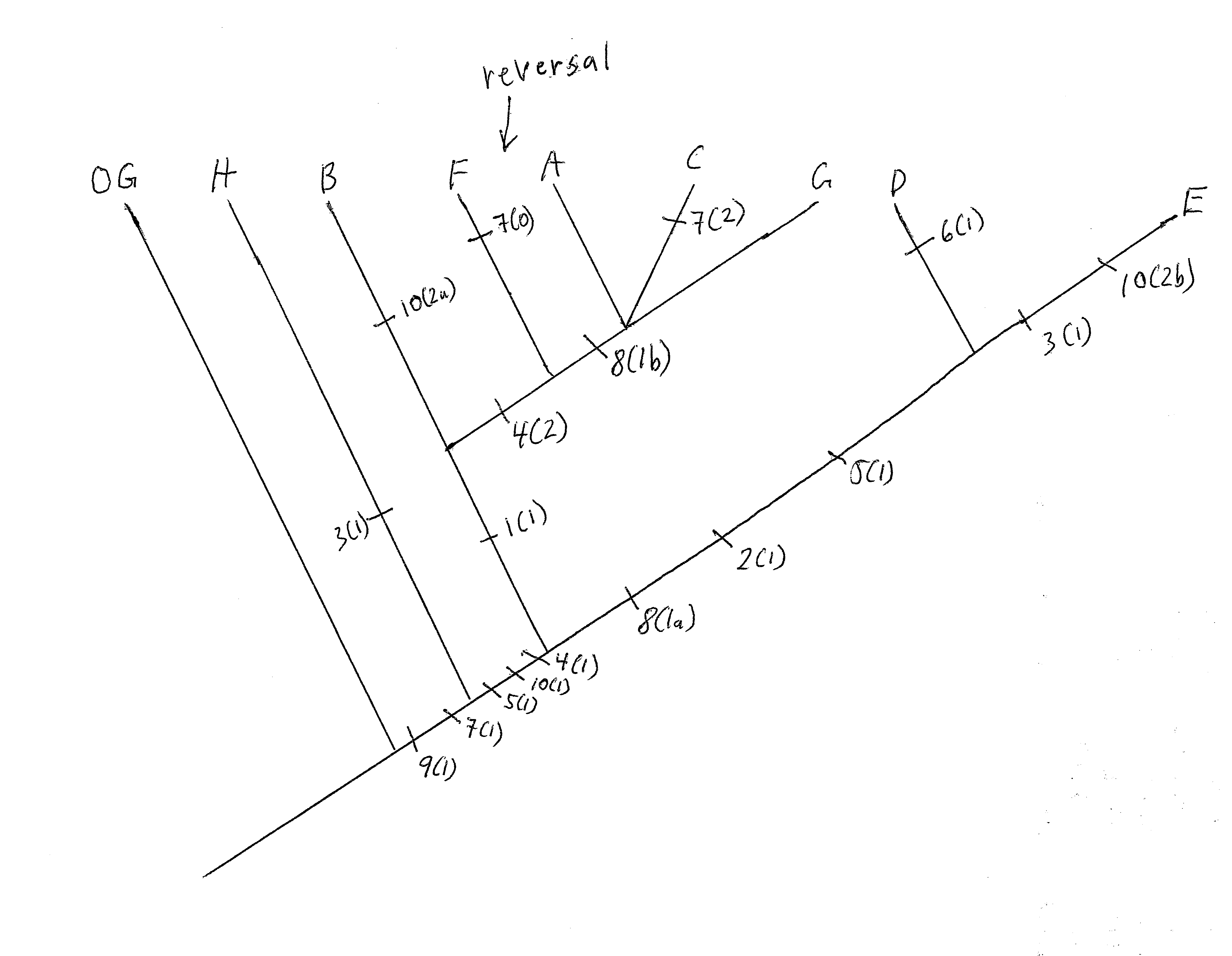
Constructing pylogenies
The Character Matrix
| Characters | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 |
| Taxon | |||||||||||
| Outgroup | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Species A | 1 | 0 | 0 | 2 | 1 | 0 | 1 | 1b | 1 | 1 | 0 |
| Species B | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 2a | 0 |
| Species C | 1 | 0 | 0 | 2 | 1 | 0 | 2 | 1b | 1 | 1 | 0 |
| Species D | 0 | 1 | 0 | 1 | 2 | 1 | 1 | 1a | 1 | 1 | 0 |
| Species E | 0 | 1 | 1 | 1 | 2 | 0 | 1 | 1a | 1 | 2b | 0 |
| Species F | 1 | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 1 | 1 | 0 |
| Species G | 1 | 0 | 0 | 2 | 1 | 0 | 1 | 1b | 1 | 1 | 0 |
| Species H | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 |
Characters
In essence, you would want to look at
as many characters as possible; you would certainly like to look at many
more character states than there are species you are trying to investigate. In
this example there are eleven characters that have been investigated. The
best characters are typically discrete traits, as continuous traits can vary and
present the investigator with a continuum (which would be divided into
separate states arbitrarily). NEVER list states such as large/small,
light/dark, long/short, etc. In order to polarize the character states,
you need an outgroup -- a species/group of species that is considered
relatively closely related that allows you to get an idea of the "ancestral"
state of the characters your are investigating, which in turn allows you to
polarize the traits.
Character States
0 : This indicates the presumptive ancestral state
1 : This is the state that is presumed to be derived from the ancestral
trait (a potential apomorphy)
1a/1b : These are different derived character states from the ancestral character
(independent of each other).
2 : This is the state that is presumed to be derived from state 1 (though
be careful about presumptions).
2a/2b: These are different derived character states from character state 1
(independent of each other).
and so on . . . .
So, understand that if species has character state "2" for a particular trait, then ancestrally that species had to have "passed through" character state "1" to get to "2". Therefore, character state "1" would unite this species with any other species that still have character state "1".
Having said that, then the following character states unite the following species:
1(1) : A, B, C, F, G
2(1) : D, E
3(1) : E, H
4(1) : A - G
4(2) :
A, C, F, G
5(1) : A - G
5(2) :
D, E
6(1) : D
7(1) : A - E, G, H (though as
you will find out shortly, species F actually has a reversal of this trait)
7(2) : C
8(1a) : D, E
8(1b) : A, C, G
9(1) : A - H (does not include the outgroup [OG])
10(1) : A - G
10(2a) :
B
10(2b) : E
11 : no relevant information available from this trait
In class construction of a phylogeny.
Analyze which species share which character states for EACH trait. The analysis should allow you to make calculations as to who is most closely related to whom and construct the tree that is most parsimonius for the group of species you are investigating. We will do this IN CLASS, with the above character matrix.
Some observations:
1. For the ingroup of 8 species, character
9 (and 7) roots the ingroup, and allow for polarization of the character states.
2. H seems to be very different from the other species, as A - G are
united by three character states [4(1), 5 (1), and 10(1)] that exclude H.
That would seem to indicate that H branches off basally on the tree, as
indicated.
3. D and E seem to be very closely related and united by 2(1),
5(2), and 8(1a).
4. Similarly, the other species (A - C and F - G) are
also united by one character, 1(1) -- so the two main branches on the tree are
[A - C, F - G] and [D & E], as indicated.
5. A, C, F, and G are united
by one character 4(2) (excluding B), resulting in that further branch on the
indicated tree, and, of those, A, C, and G are further united by one character
8(1b) (excluding F), which results in the final branch as indicated.

Any unresolved polytomies?
Any reversals?
Any independent derivations of the same trait?
Any species not supported by an autapomorphy (from the data set)?
How many genera supported by the data set (you will need the phylogeny to tell)?
A couple more character matrices to practice with:
|
Taxon \ Character |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
|
Outgroup |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
|
Species A |
1 |
1 |
0 |
2 |
1 |
0 |
1a |
|
Species B |
2 |
2 |
1 |
0 |
2b |
0 |
0 |
|
Species C |
1 |
1 |
0 |
1 |
1 |
0 |
0 |
|
Species D |
2 |
1 |
1 |
0 |
1 |
1 |
0 |
|
Species E |
2 |
2 |
1 |
0 |
2b |
0 |
1b |
|
Species F |
1 |
1 |
0 |
2 |
2a |
0 |
0 |
2.
|
Taxon \ Character |
1 |
2 |
3 |
4 |
5 |
6 |
7 |
|
Outgroup |
0 |
0 |
0 |
0 |
0 |
0 |
0 |
|
Species A |
0 |
0 |
1 |
0 |
2b |
0 |
0 |
|
Species B |
1 |
1 |
2 |
1 |
1 |
2 |
0 |
|
Species C |
0 |
1 |
1 |
2 |
1 |
0 |
1 |
|
Species D |
1 |
1 |
2 |
1 |
1 |
1 |
0 |
|
Species E |
0 |
2 |
1 |
2 |
2a |
0 |
1 |
|
Species F |
0 |
2 |
1 |
2 |
2a |
0 |
1 |